
Jeanne Ropars
Position: Research Scientist
Detailed position: Research Scientist CNRS
Team: Evolutionary genetics and ecology
Contact details:
Laboratoire Écologie, Société et Évolution – IDEEV
Bureau 2316 Bât. 680 – 12, route 128
91190 Gif Sur Yvette
Tel: +33 (0)1 69 15 46 93
Fax: –
Email : jeanne.ropars (at) universite-paris-saclay.fr
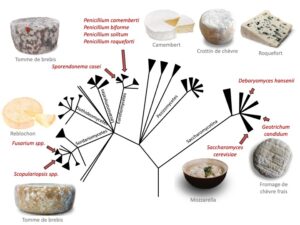
Is adaptation repeatable?
Research interests
I am interested in the mechanisms involved in adaptation, including horizontal gene transfer, sexual reproduction, gene flow and genetic diversity. I think that fungi are good models for testing theories and developing new concepts in a unique way. Indeed, fungi are excellent models for studies of evolution and adaptation in eukaryotes, given their many experimental advantages. Some fungi have small genomes, are easy to culture in laboratory conditions, have short generation times, remain alive for long periods in the freezer, are easy to transform genetically and have a rich diversity of lineages and ecological niches.
These characteristics make it possible to address evolutionary biology questions with combinations of complementary approaches, including genomic and experimental methods. In particular, the use of multiple, distantly related lineages, of fungi in the production of certain types of food creates an ideal model system for studying the repeatability of evolution, by providing several independent recent replicates of adaptation to the same ecological niche under strong and recent selection. Since 2018, I am studying the repeatability of evolution using cheese fungi as models, based on genome analyses, historical data and experiments.
Image: My research project: is adaptation repeatable ? Cheese fungi are ideal models to study parallel adaptation because 1) several phylogenetically distant lineages are found in the same ecological niche and 2) this adaptation is the result of strong and recent selection (< 8000 years). My research questions are: Did these parallel adaptations lead to convergent phenotypes? Are the same genes/genomic regions and the same genomic mechanisms involved in these parallel adaptations? Tree modified from Lifemap (http://lifemap.univ-lyon1.fr/).
Career path
2022 : Habilitation à Diriger des Recherches, University of Paris-Saclay. Defended on June 30, 2022
2018- : Permanent CNRS researcher position (Chargée de recherche CNRS), University of Paris-Sud XI, Orsay
2016-2018 : Post-Doc, Institut Pasteur, Paris, France – C. d’Enfert’s group, working on the human fungal pathogen Candida albicans
2014-2015 :Post-Doc, University of Ottawa, Ottawa, Canada – N. Corradi’s group, working on arbuscular mycorrhizal fungi
2012-2014 : Post-Doc, University of Paris-Sud XI, Orsay, France – T. Giraud’s group, working on Penicillium cheese fungi
2009-2012 : PhD in evolutionary biology, National Museum of Natural History, Paris, France – Supervision J. Dupont. Title: “Systematics, adaptive mechanisms and sexual reproduction in cheese fungi”. Defended on June 22, 2012
2006-2008 : Master in Systematics and Evolution, National Museum of Natural History, Paris, France
Publications
2022
Ropars J, Giraud T. Convergence in domesticated fungi used for cheese and dry-cured meat maturation: beneficial traits, genomic mechanisms and degeneration. Current Opinion in Microbiology. In Press
Lo Y-C, Bruxaux J, Vega RCR de la, Snirc A, Coton M, Piver ML, et al. Footprints of domestication in dry-cured meat Penicillium fungi: convergent specific phenotypes and horizontal gene transfers. bioRxiv. 2022. p. 2022.03.25.485132. https://www.biorxiv.org/content/10.1101/2022.03.25.485132v1.full.pdf
Bennetot B, Vernadet J-P, Perkins V, Hautefeuille S, Rodriguez de la Vega RC, O’Donnell S, et al. Domestication of different varieties in the cheese-making fungus Geotrichum candidum. bioRxiv. 2022;2022.05.17.492043. https://www.biorxiv.org/content/10.1101/2022.05.17.492043v3.full.pdf
2021
Savary O, Coton M, Frisvad J, Nodet P, Ropars J, Coton E, et al. Unexpected Nectriaceae species diversity in cheese, description of Bisifusarium allantoides sp. nov., Bisifusarium penicilloides sp. nov., Longinectria gen. nov. lagenoides sp. nov. and Longinectria verticilliforme sp. nov. Mycosphere. 2021;12:1077–100. https://www.mycosphere.org/pdf/MYCOSPHERE_12_1_13-1.pdf
2020
Ropars J, Didiot E, Rodríguez de la Vega RC, Bennetot B, Coton M, Poirier E, et al. Domestication of the emblematic white cheese-making fungus Penicillium camemberti and its diversification into two varieties. Current Biology. 2020;30:4441–53. https://www.cell.com/current-biology/fulltext/S0960-9822(20)31272-0.
Ropars J, Caron T, Lo Y, Bennetot B, Giraud T. The domestication of Penicillium cheese fungi. Comptes Rendus Biologies. 2020;343:155–76. https://comptes-rendus.academie-sciences.fr/biologies/article/CRBIOL_2020__343_2_155_0.pdf
Hernández-Cervantes A, Znaidi S, van Wijlick L, Denega I, Basso V, Ropars J, et al. A conserved regulator controls asexual sporulation in the fungal pathogen Candida albicans. Nature Communications. 2020;11:6224. https://www.nature.com/articles/s41467-020-20010-9
Dumas E, Feurtey A, Vega RCR de la, Prieur SL, Snirc A, Coton M, et al. Independent domestication events in the blue-cheese fungus Penicillium roqueforti. Molecular Ecology. 2020;29:2639–60. https://onlinelibrary.wiley.com/doi/full/10.1111/mec.15359
Chen ECH, Mathieu S, Hoffrichter A, Ropars J, Dreissig S, Fuchs J, et al. More Filtering on SNP Calling Does Not Remove Evidence of Inter-Nucleus Recombination in Dikaryotic Arbuscular Mycorrhizal Fungi. Front Plant Sci 1. https://www.frontiersin.org/articles/10.3389/fpls.2020.00912/full?&utm_source=Email_to_authors_&utm_medium=Email&utm_content=T1_11.5e1_author&utm_campaign=Email_publication&field=&journalName=Frontiers_in_Plant_Science&id=543611#B2
2018
Ropars J, Maufrais C, Diogo D, Marcet-Houben M, Perin A, Sertour N, et al. Gene flow contributes to diversification of the major fungal pathogen Candida albicans. Nature Communications. 2018;9:2253. https://www.nature.com/articles/s41467-018-04787-4
Chen EC, Mathieu S, Hoffrichter A, Sedzielewska-Toro K, Peart M, Pelin A, et al. Single nucleus sequencing reveals evidence of inter-nucleus recombination in arbuscular mycorrhizal fungi. Wittkopp PJ, Martin F, Geurts R, Young P, Seidl M, editors. eLife. 2018;7:e39813.
Chen ECH, Morin E, Beaudet D, Noel J, Yildirir G, Ndikumana S, et al. High intraspecific genome diversity in the model arbuscular mycorrhizal symbiont Rhizophagus irregularis. New Phytologist. 2018;220:1161–71.
2017
Ropars J, López-Villavicencio M, Snirc A, Lacoste S, Giraud T. Blue cheese-making has shaped the population genetic structure of the mould Penicillium roqueforti. PLOS ONE. 2017;12:e0171387.
Dupont J, Dequin S, Giraud T, Le Tacon F, Marsit S, Ropars J, et al. Fungi as a Source of Food. Microbiology Spectrum. 2017; 5
d’Enfert C, Bougnoux M-E, Feri A, Legrand M, Loll-Krippleber R, Marton T, et al. Genome Diversity and Dynamics in Candida albicans. 2017; Pp. 205–232 in R. Prasad, ed. Candida albicans: Cellular and Molecular Biology. Springer International Publishing, Cham.
2016
Ropars J, Toro KS, Noel J, Pelin A, Charron P, Farinelli L, et al. Evidence for the sexual origin of heterokaryosis in arbuscular mycorrhizal fungi. Nature Microbiology. 2016;16033.
Ropars J, Lo Y, Dumas E, Snirc A, Begerow D, Rollnik T, et al. Fertility depression among cheese‐making Penicillium roqueforti strains suggests degeneration during domestication. Evolution. 2016;70:2099–109.
Ropars J, de la Vega RCR, López-Villavicencio M, Gouzy J, Dupont J, Swennen D, et al. Diversity and Mechanisms of Genomic Adaptation in Penicillium. Aspergillus and Penicillium in the Post-genomic Era. 2016; P. 210 in Aspergillus and Penicillium in the Post-genomic Era. Ronald P. de Vries, Isabelle Benoit Gelber, Mikael Rordam Andersen.
2015
Ropars J, Rodríguez de la Vega RC, López-Villavicencio M, Gouzy J, Sallet E, Dumas É, et al. Adaptive horizontal gene transfers between multiple cheese-associated Fungi. Current Biology. 2015;25:2562–9. https://www.cell.com/current-biology/pdfExtended/S0960-9822(15)00996-3
Ropars J, Corradi N. Homokaryotic vs heterokaryotic mycelium in arbuscular mycorrhizal fungi: different techniques, different results? New Phytologist. 2015;208:638–41.
Pintye A, Ropars J, Harvey N, Shin H-D, Leyronas C, Nicot PC, et al. Host Phenology and Geography as Drivers of Differentiation in Generalist Fungal Mycoparasites. Zhang Z, editor. PLOS ONE. 2015;10:e0120703.
Gillot G, Jany J-L, Coton M, Le Floch G, Debaets S, Ropars J, et al. Insights into Penicillium roqueforti morphological and genetic diversity. Lumbsch HT, editor. PLOS ONE. 2015;10:e0129849.
Cornille A, Feurtey A, Gélin U, Ropars J, Misvanderbrugge K, Gladieux P, et al. Anthropogenic and natural drivers of gene flow in a temperate wild fruit tree: a basis for conservation and breeding programs in apples. Evolutionary Applications. 2015;8:373–84.
2014
Ropars J, López-Villavicencio M, Dupont J, Snirc A, Gillot G, Coton M, et al. Induction of sexual reproduction and genetic diversity in the cheese fungus Penicillium roqueforti. Evolutionary Applications. 2014;7:433–41.
Ropars J, Aguileta G, de Vienne DM, Giraud T. Massive gene swamping among cheese-making Penicillium fungi. Microbial Cell. 2014;1.
Gladieux P, Ropars J, Badouin H, Branca A, Aguileta G, de Vienne DM, et al. Fungal evolutionary genomics provides insight into the mechanisms of adaptive divergence in eukaryotes. Molecular Ecology. 2014;23:753–73.
Cheeseman K, Ropars J, Renault P, Dupont J, Gouzy J, Branca A, et al. Multiple recent horizontal transfers of a large genomic region in cheese making fungi. Nat Commun. 2014;5:2876.
2012
Ropars J, Dupont J, Fontanillas E, Rodríguez de la Vega RC, Malagnac F, Coton M, et al. Sex in cheese: evidence for sexuality in the fungus Penicillium roqueforti. Corradi N, editor. PLoS ONE. 2012;7:e49665.
Ropars J, Cruaud C, Lacoste S, Dupont J. A taxonomic and ecological overview of cheese fungi. International Journal of Food Microbiology. 2012;155:199–210.
2010
Giraud F, Giraud T, Aguileta G, Fournier E, Samson R, Cruaud C, et al. Microsatellite loci to recognize species for the cheese starter and contaminating strains associated with cheese manufacturing. International Journal of Food Microbiology. 2010;137:204–13.
